-
CBI alumnus Yadilette Rivera-Colon wins award!
We are proud to announce that Yadilette Rivera-Colon is one of nine winners of the Women of Color STEM Achievement Awards foe 2021. See the full announcement here. Yadilette completed her PhD in the Garman Lab in 2013, and is now an Assistant Professor of Biology at Bay Path University.
-
Chalk Talk May 5, 2021 – CBI Trainees
“Force-responsive Materials Utilizing Cryptic Crosslinking Sites“Adrian Lorenzana (Peyton Lab) “Establishing Adhesive Responses of Uropathogenic E. coli to Gels with Various Stiffnesses“Brandon Barajas (Schiffman Lab) “Metabolic Labeling as a Discovery Tool for Cell Wall Recycling“Rebecca Gordon (Siegrist Lab) “Chemical labeling of Mycobacteria for growth monitoring and detection“Kiserian Jackson (Siegrist Lab) Please see our full dates for…
-
Chalk Talk April 7, 2021 – Min Chen & Vince Rotello
“Direct Cytosolic Delivery of GFP and the Awesome Power of Nuclear Fluorescence“David Luther (Rotello Lab) “Development of a Multiplex OmpG Biosensor“Josh Foster (M. Chen Lab) “Studying the conformational dynamics of Abl kinase in real time with a nanopore tweezer“Fanjun Li (M. Chen Lab) Please see our full dates for Fall 2020-Spring 2o21 and presenting labs…
-
Chalk Talk March 3, 2021 – Yasu Morita and Dan Hebert
“Polyprenol biosynthesis in the maintenance and regulation of mycobacterial membrane domain“Student Speaker -Malavika Prithviraj (Yasu Lab ) “Understanding the selectivity of the UGGTs, key quality control sensors and gatekeepers of the mammalian secretory pathway“Student Speaker – Kevin Guay (Hebert Lab) “The identification and characterization of a putative ER adapter protein TTC17“Student Speaker – Nathan Canniff…
-
CBI Students’ Publication: Ben Adams, Nathan Canniff and Kevin Guay reveal the specificity of key protein quality control sensors
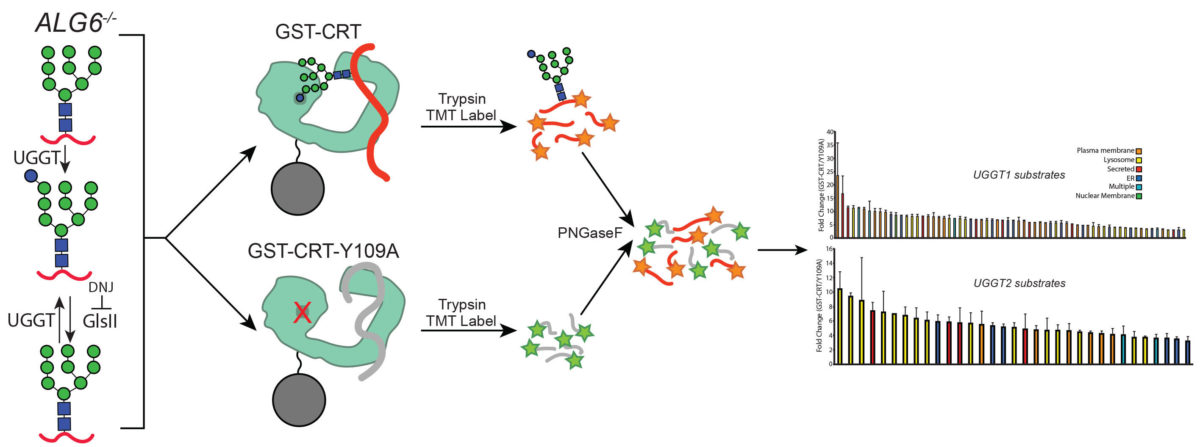
UGGT1 and UGGT2 are key quality control factors that determine the fate of glycoproteins in the early mammalian secretory pathway. These two paralogues direct persistent molecular chaperone binding in the endoplasmic reticulum that helps with protein maturation and sorting. Persistent chaperone binding of terminally misfolded clients can target proteins for degradation by the proteasome, as…
-
Student Publication: Carey Dougan, “Cavitation in Soft Matter” 2020
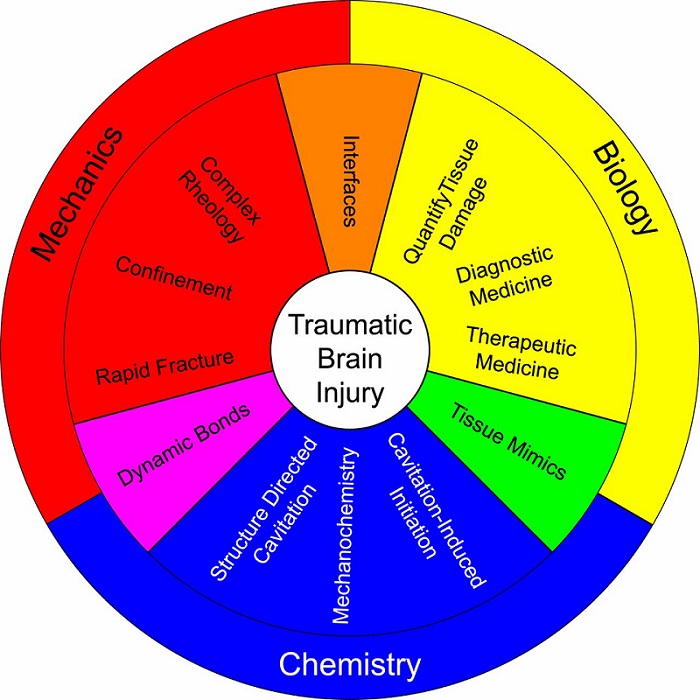
Co-first author Dougan adds, “While cavitation is often thought of as something to be avoided, we aim to use it to benefit medicine and the development of new treatments.” For example, cavitation rheology can be used to measure the strength of interfaces within the brain, which is difficult to achieve with any other method, she…
-
Weiyue Xin – Graduate Research Student Seminar (GRASS) 3rd Place
CBI Trainee Weiyue Xin (Santore Lab) took third place in the Annual G.R.A.S.S. (Graduate Research Student Symposium) for her seminar on the interactions between solid domains in biomimetic membranes. Weiyue’s work is part of a larger DOE-sponsored collaboration between the Santore and Grason groups, aiming to understand new mechanisms occurring in membranes made from biomolecules,…
-
Chalk Talk Feb 3, 2021 – Lynmarie Thompson and Richard Vachet
“Investigating E. coli CheA Activation by Chemoreceptors using Hydrogen Deuterium Exchange Mass Spectrometry”Student Speaker – Thomas Tran (Thompson Lab) “DNA-Mediated Assembly of Functional Chemoreceptors: Using Structural DNA Nanotechnology to Advance Biophysical Research” Student Speaker – Tiernan Kenneday (Thompson Lab) “Membrane Protein Structure and Binding Interactions Studied by Mass Spectrometry” Student Speaker – Xiao Pan (Vachet…
-
Chalk Talk Dec 2, 2020 – S. Thai Thayumanavan and Patrick Flaherty Labs
“Estimating Subclonal Populations from Structured DNA Sequencing Data“Student Speaker – Shai He (Flaherty Lab) “Antibody – Nanoparticle Conjugates for Targeted Delivery of Chemotherapeutics“Student Speaker – Khushboo Singh (Thayumanavan Lab)
-
Chalk Talk Nov 4, 2020 – Eric Strieter and Mingxu You Labs
“A Cryptic Site on the Proteasome-Associated Deubiquitinase UCH37/UCHL5 is Required for Chain Editing and Degradation“Student Speaker – Jiale Du (Strieter Lab) “Real-time Monitoring of Transient Lipid-Lipid Interactions in Live Cell Membranes using a DNA Prob“Student Speaker – Yousef Bagheri Zarringhabae (You Lab) “Multiplex Imaging in Living Cells with Fluorogenic RNAs“Student Speaker – Ru Zheng (You…
-
Chalk Talk Oct 7, 2020 – Jeanne Hardy and Amanda Woerman Labs
“Studying allosteric sites on caspase-6 to aid the development of substrate-specific inhibitors“Student Speaker: Irina Sagarbarria and Andrew J. Smith (Hardy Lab) “Investigating alpha-synuclein aggregation using novel models of strain competition“Student Speaker: Sarah Holec (Woerman Lab)